Transcriptomic Analysis of Quinoa Reveals a Group of Germin-Like Proteins Induced by Trichoderma
Symbiotic strains of fungi in the genus Trichoderma affect growth and pathogen
resistance of many plant species, but the interaction is not known in molecular
detail. Here we describe the transcriptomic response of two cultivars of the crop
Chenopodium quinoa to axenic co-cultivation with Trichoderma harzianum BOL-12
and Trichoderma afroharzianum T22. The response of C. quinoa roots to BOL-12 and
T22 in the early phases of interaction was studied by RNA sequencing and RT-qPCR
verification. Interaction with the two fungal strains induced partially overlapping gene
expression responses. Comparing the two plant genotypes, a broad spectrum of putative
quinoa defense genes were found activated in the cultivar Kurmi but not in the Real
cultivar. In cultivar Kurmi, relatively small effects were observed for classical pathogen
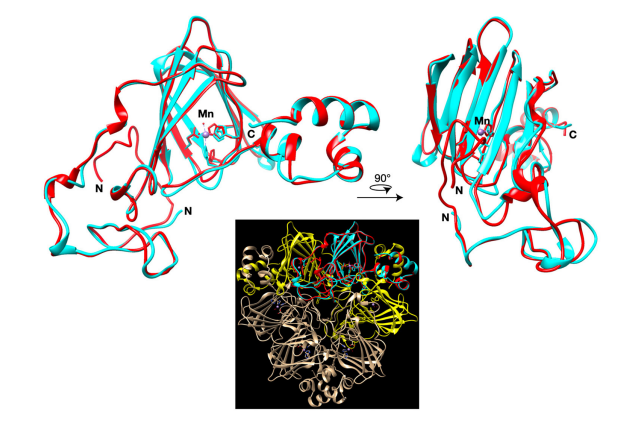
response pathways but instead a C. quinoa-specific clade of germin-like genes were
activated. Germin-like genes were found to be more rapidly induced in cultivar Kurmi
as compared to Real. The same germin-like genes were found to also be upregulated
systemically in the leaves. No strong correlation was observed between any of the known
hormone-mediated defense response pathways and any of the quinoa-Trichoderma
interactions. The differences in responses are relevant for the capabilities of applying
Trichoderma agents for crop protection of different cultivars of C. quinoa.
https://www.frontiersin.org/articles/10.3389/ffunb.2021.768648/full